Running Batch with Nvidia Clara Parabricks
Thomas Leung
HPC Technical Solution Consultant, Professional Service Organization, Google Cloud
Try Google Cloud
Start building on Google Cloud with $300 in free credits and 20+ always free products.
Free trialRunning a lot of genomic pipelines can be challenging. Adding GPUs to the solution is harder. In this blog, we are presenting the whole solution of running multiple jobs with GPU and Batch. As Nvidia Clara Parabricks is available in the Google Cloud Platform Marketplace, you can easily spin up a single VM with Parabricks tools to help with your genomics pipeline workloads. A Parabricks VM allows you to run individual workflow(s) manually. If we are in a Lab / Health Care institute level environment, a scale-out solution with deployment automation is needed to run analysis in parallel. Creating multiple Parabricks VMs is not the solution either because there is no central job management across VMs created from the Marketplace. Batch is the solution addressing the problem. A lot of Genomic pipelines rely on the HPC solutions and toolings like Parabricks. We are going to discuss how and why to run Nvidia Clara Prarbricks on Batch at scale.
Product Summary:
Batch
Fully managed batch service to schedule, queue, and execute batch jobs on Google's infrastructure. User provisions and auto scales capacity while eliminating the need to manage third-party solutions. It is natively integrated with Google Cloud to run, scale, and monitor workload.
Nvidia Clara Parabricks Pipelines
Parabricks is a software suite for performing secondary analysis of next generation sequencing (NGS) DNA and RNA data. A major benefit of Parabricks is that it is designed to deliver results at blazing fast speeds and low cost. Parabricks can analyze whole human genomes in about 45 minutes, compared to about 30 hours for 30x WGS data. The best part is the output results exactly match the commonly used software. So, it’s fairly simple to verify the accuracy of the output.
Processing flow:
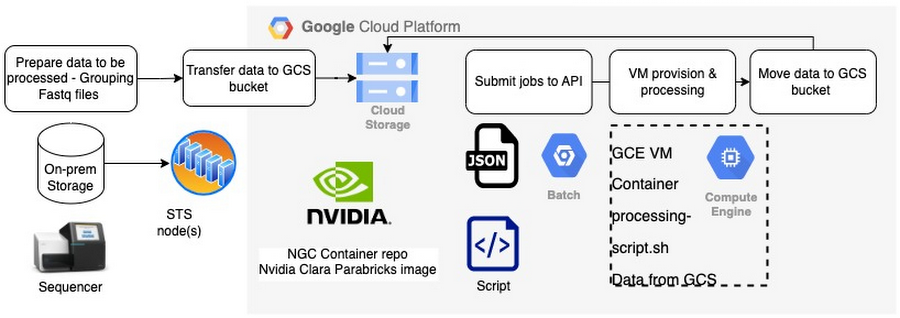

Provided the data is ready in the Google Cloud Storage bucket, you can submit jobs to the Batch API,
All you have to worry about is 2 files.
Batch json file - This file defines everything about the infrastructure including machine type, # of GPU, container image, persistent disk, GCS bucket (data) location and run script location and commands etc.
Bash script runs within the docker container - This file has all steps to be run.
Make sure the Batch API is enabled and you have the IAM roles / permissions to submit the job.
In the following example, we are going to follow the tutorial steps from Nvidia Clara Parabricks website:
https://docs.nvidia.com/clara/parabricks/3.8.0/Tutorials.html
Architecture and Infrastructure decisions:
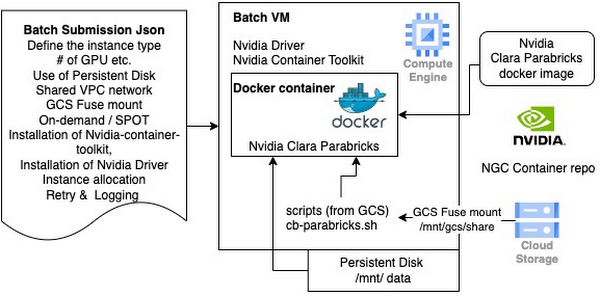

Use the Compute Engine Persistent Disk - users can define the size and the performance. Size can be up to 63TB per disk. Performance tier between pd-standard , pd-balanced, pd-ssd.
Use Google Cloud Storage (GCS)
Scripts can be mounted to the Batch VM and the Docker container.
Input and Output can be stored cost effectively in GCS buckets
For large datasets, transfer operations like GSUTIL may perform better.
Nvidia Clara Parabricks image - use the Parabricks image natively without building a new image. You don’t need to maintain a custom image.
Sample code:
Here is the sample GCP environment summary which includes shared VPC network use case:
GCP service project: service-hpc-project2
GCP shared VPC host project: host-hpc-project1
GCP network: test-network
GCP subnetwork: tier-1
GCS Bucket: thomashk-test2
Batch Submission Json example (cb-parabricks.json)
#Copyright 2023 Google. This software is provided as-is, without warranty or representation for any use or purpose. Your use of it is subject to your agreement with Google
This is the script with running steps with a batch file (parabricks-run.sh)
Store the script in the GCS bucket (gs://thomashk-test2)
$ gsutil ls gs://thomashk-test2/parabricks
gs://thomashk-test2/parabricks/
gs://thomashk-test2/parabricks/parabricks-run.sh
gs://thomashk-test2/parabricks/input/
gs://thomashk-test2/parabricks/output/
User can submit the job with the following command:
Job submission command:
Job status checking command:
With Batch and Parabricks, we can accelerate genomic runs with GPUs and operate at scale. You may consider generating genomic runs scripts as data available at the storage and cron jobs to submit jobs into Batch automatically without any interactions. Batch offers status of the jobs via API, gcloud command and webGUI.
Happy computing!